제품설명
Ovation Ultralow Methyl-Seq Library Systems은 DNA methylation 분석을 위해 bisulfite sequencing에 사용되는 library를 제작하는 간편하고 빠른 방법이다. 이 시스템은 10 ng의 DNA
정도만 필요하기 때문에 다양한 샘플의 methylation 연구에 적용 가능하다. 프로토콜은 whole genome bisulfite sequencing과 호환 가능하고 bisulfite conversion 단계를 포함해 대략
9시간 정도면 실험을 완료할 수 있다.
Figure 1에서 보는 것과 같이 genomic DNA의 fragmentation, blunt ends 생성을 위한 end repair, adaptor ligation,그리고 5-methyl-cytosine dNTPs로 fill-in이라는 4개의 주요 단계로
구성되어 있다. Adaptor ligation 다음에 bisulfite conversion이 수행된다.
Ovation Ultralow Methyl-Seq Library Systems은 Illumina sequencing platforms에서 multiplexing이 가능하도록 두개의 kit 형태로 되어 있다. Ultralow Methyl-Seq DR Multiplex
System 1-8 (Part No. 0335)과 Ovation Ultralow Methyl-Seq DR Multiplex System 9-16 (Part No. 0336)는 각각 8개의 unique barcoded adaptors로 multiplex sequencing이 가능하며
이 kit를 조합하여 사용하면 16-plex sequencing까지 가능하다.
Figure 1. The Ovation Ultralow Methyl-Seq Library System Workflow
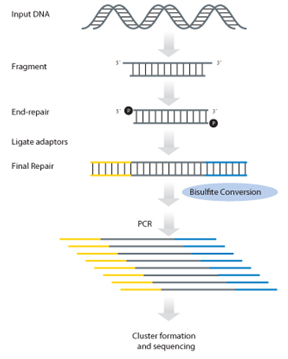
Figure 2. Coverage uniformity
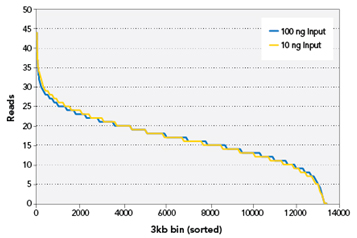
Figure 2. Coverage uniformity. Whole genome bisulfite sequencing was performed on libraries generated from 100ng and 10ng of human genomic DNA.
Sequencing was performed on the Illumina GAIIx platform. From each library, 225,000 reads that mapped to a 40 million bp region of Chromosome 5 were randomly
selected. This region was divided into 3 kbp bins. The number of reads with a start position within that bin was recorded. Bins were then sorted by number of
reads from high to low and plotted.
Figure 3. Methylated CpG quantification
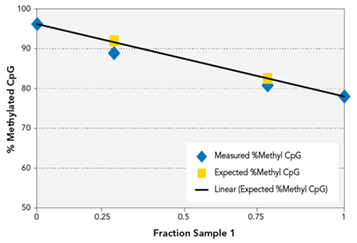
Figure 3. Methylated CpG quantification. Libraries were made from 10ng Sample 1 (Normal Human gDNA), 10ng Sample 2 (in vitro CpG methylated
Human gDNA), 10ng of a 25:75 mixture of Sample 1:Sample 2, and 10ng of a 75:25 mixture. Libraries were sequenced to determine the % Methyl CpG present on average
across all reads uniquely mapping to the genome. Sample 1 and Sample 2 % Methyl CpG values were used to calculate the expected % Methyl CpG present in the mixtures.



