※ 변경사항
본 제품의 제품명이 변경되었습니다. (2026년 기준)
(기존) SMART-Seq® v4 3’ DE Kit
(변경) SMART-Seq® mRNA 3' DE
자세한 내용은 다카라코리아 (02-2081-2510)로 문의 주세요.
- SMART-Seq® v4 기반으로 single-cell 또는 10pg의 극소량 total RNA 적용
- 전사체의 3’ 말단만 특이적으로 분석하여 합리적인 가격으로 High-throughput Differential Expression (DE) 분석 가능
- 최대 12 Multiplexing Illumina® Library 제작 가능
최상의 결과를 보장하는 SMART 기술
단일세포 (Single cell) 전사체 분석은 ‘세포집단 (cell population) 내에서 각각의 세포의 어떤 유전자 발현율을 보이는지’ 를 이해하는 데
중요하다. SMART-Seq® v4 3’ DE Kit는 단일세포 유래의 mRNA의 3’ 말단의 서열 정보를 얻을 수 있어 유전자 발현차이 분석 (Differential gene Expression (DE)
analysis)에 최적이다. 본 제품은 SMART®법과 Locked Nucleic Acid (LNA) 기술을 응용한 SMART-Seq® v4 키트의 특징을 도입하였다.
mRNA의 3’ 말단만 집중적으로 서열을 분석 가능하며, 본 제품 내 Read2의 바코드로 인식, 사용할 수 있는 In-line 인덱스가 포함되기 때문에 샘플까지
Multiplexing NGS Library를 제작 및 분석이 가능하다. 1 ~ 10.5ul의 RNA volume을 적용 가능하며 제작된 cDNA는 Illumina Nextera® XT DNA Library
Preparation Kit*1를 이용하여 NGS Library 제작*2이 가능하다. 본 제품을 사용하여
제작된 NGS Library는 Illumina® HiSeq®, NextSeq®*3, MiSeq®, and MiniSeq™*3에
적용할 수 있다.
*1: Fragmentation과 Tagging 과정을 위해 Nextera® XT DNA Library Preparation Kit가 별도 필요
*2: Library index는 이론적으로 최대 1,152 (=12 x 96)개까지 가능
*3: NextSeq®, MiniSeq™ 시퀀서로 시퀀싱분석을 적용할 경우, 샘플에 Control DNA (PhiX) 추가 필요
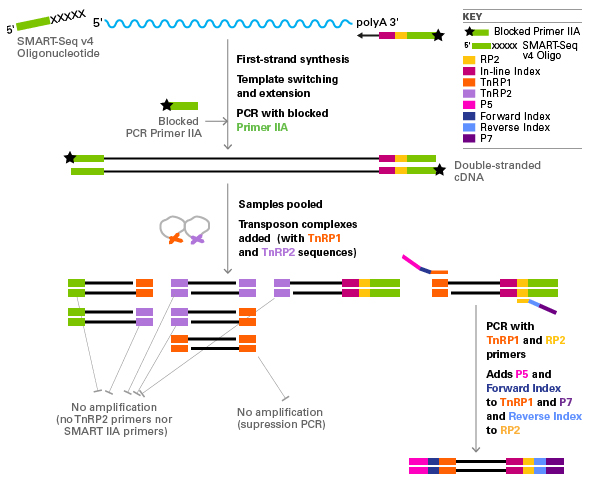
그림 1. SMART-Seq® v4 3' DE Kit를 이용한 cDNA 합성과 Library 제작
cDNA (black) is synthesized with a blocked (black star) and modified oligo(dT) primer that adds sequences for subsequent amplification and
analysis-an in-line index (magenta), part of the Illumina read primer 2 sequence (RP2, yellow), and the SMART IIA sequence (green).
The SMART IIA sequence is used as a priming site during cDNA amplification, the Illumina RP2 sequence is used as a priming site during library
amplification, and the in-line index is used for demultiplexing pooled samples during analysis. The process works as follows: first, the template
for SMARTScribe reverse transcriptase switches from the mRNA (blue wavy line) to the SMART-Seq v4 Oligonucleotide (green). After reverse transcription,
the full-length cDNA is amplified by PCR with blocked Primer IIA oligonucleotides. After cDNA amplification, the presence of the in-line index (magenta)
allows for pooling of up to 12 samples. The pooled samples are tagmented and Illumina Nextera read primer 1 and 2 sequences are added by the
Nextera Tn5 transposon (TnRP1 and TnRP2, orange and purple respectively). The 3' ends of the original mRNA are captured by selective PCR with primers
for the TnRP1 and RP2 sequences. Other products of the transposon-based reaction are not amplified, either because there are no primer sites for
amplification or because of suppression PCR. Cluster generation (pink and dark purple) and indexing sequences (light blue and dark blue) are added
during this PCR stage to generate a library ready for sequencing on an Illumina platform.
Sample ID |
Pool |
In-line Indexes |
i1 |
i2 |
i3 |
i4 |
i5 |
i6 |
i7 |
i8 |
i9 |
i10 |
i11 |
i12 |
mtRNA(%) |
3.3 |
3.1 |
3.6 |
1.7 |
3.9 |
4.1 |
1.8 |
2.5 |
2.8 |
5.5 |
4.1 |
2.7 |
4.8 |
rRNA(%) |
0.6 |
1.2 |
0.7 |
1.1 |
0.4 |
0.4 |
0.3 |
0.3 |
0.7 |
1.2 |
0.5 |
0.1 |
0.6 |
Uniquely mapped reads(%) |
69 |
68 |
68 |
70 |
68 |
71 |
71 |
72 |
68 |
69 |
69 |
74 |
69 |
Total mapped reads(%) |
97 |
96 |
98 |
97 |
97 |
98 |
98 |
98 |
96 |
97 |
96 |
98 |
98 |
Number of reads(M) |
21.6 |
2.3 |
5 |
1.3 |
2.2 |
2 |
1.9 |
0.8 |
1.3 |
0.6 |
0.2 |
1.5 |
1.6 |
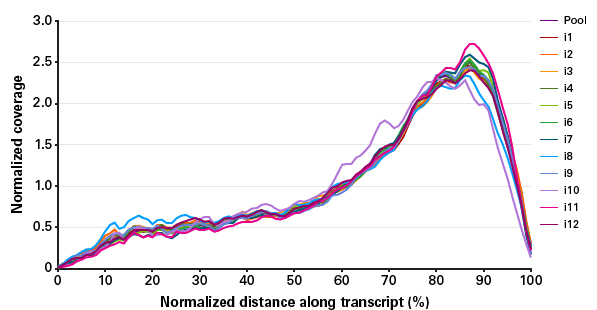
표 1. K562 single cell로부터 제작된 pooled library의 mapping read (%)
K562 cells were diluted to one cell/μl in PBS buffer and twelve single cells were isolated, checked via optical microscopy, lysed,
and subjected to cDNA synthesis. The pooled libraries were sequenced on an Illumina MiSeq instrument with 47 bp for read 1 and 26 bp
for read 2. The pooled libraries were demultiplexed based on the in-line barcode sequence from read 2. All libraries were mapped with
STAR v.2.3.0.1 (Dobin et al. 2013) against the human genome (hg19). The reads map to the genome at a high rate (>96%) with a small
proportion mapping to rRNA or mitochondrial (mt) regions.
Applications
cDNA synthesis for end-capture mRNA-seq for differential expression analysis of single cells.
구성품 (자세한 내용은 CoA를 참조하세요)
SMART-Seq v4 3' DE Kit Components
SeqAmp™ DNA Polymerase